Beta Status
Genotypic data management is under active development. BMS has a repository for SNP markers, molecular maps, and low-density genotypic data sets. There is also the ability to create samples and sample ids from plants within plots.
Data Size Capability
The BMS supports marker-based selection decisions with a low-density genotype data repository. The ability of the BMS to upload and retrieve datasets is limited somewhat by server specifications. Preliminary testing indicates the BMS installed on AWS t2.medium, can reliably upload and retrieve datasets with a maximum of 400,000 data points (100 germplasm genotyped with 4000 markers).
Upcoming
Expect to see the following improvements in upcoming releases:
- Improved and more user friendly user interface
- Improved connectivity to germplasm details and management
- Expanded sample and marker list management
- Creation of custom genotyping templates for service providers
- Repository and retrieval functionality for larger SNP datasets
- API interoperability with BrAPI enabled genotypic databases (i.e: GOBII; Gigwa) and associated tools
- Marker based summaries presented with phenotypic data to assist selection
Video: Marker-Assisted Backcrossing v9
Markers
Upload
SNP markers must be defined in the system before associated genotypic data can be uploaded. To define SNP markers, a Excel template is downloaded from the GDMS, the marker definitions are entered into the template, and the resulting marker definitions are loaded into the GDMS
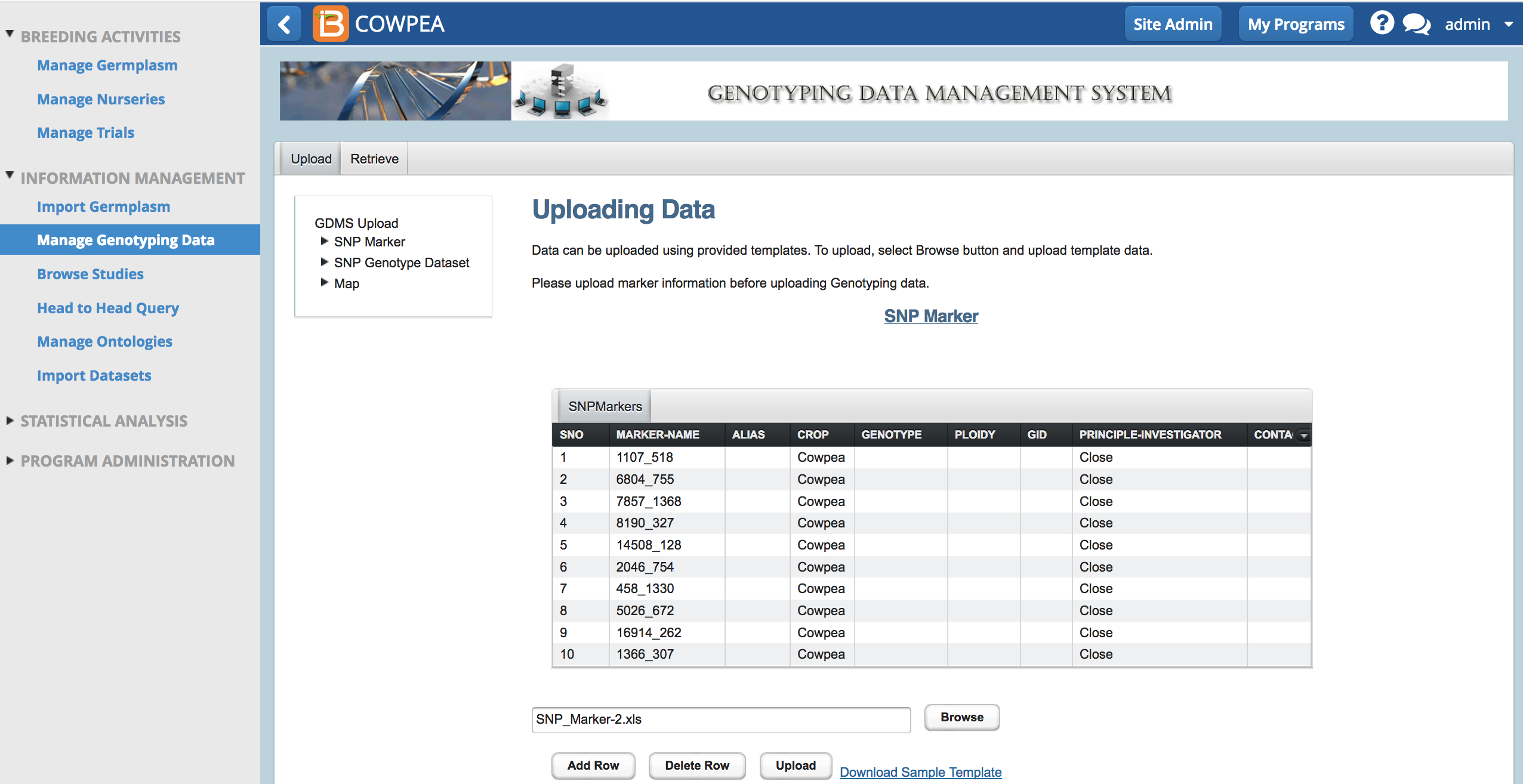
- Download marker template (.xls) from Upload tab.
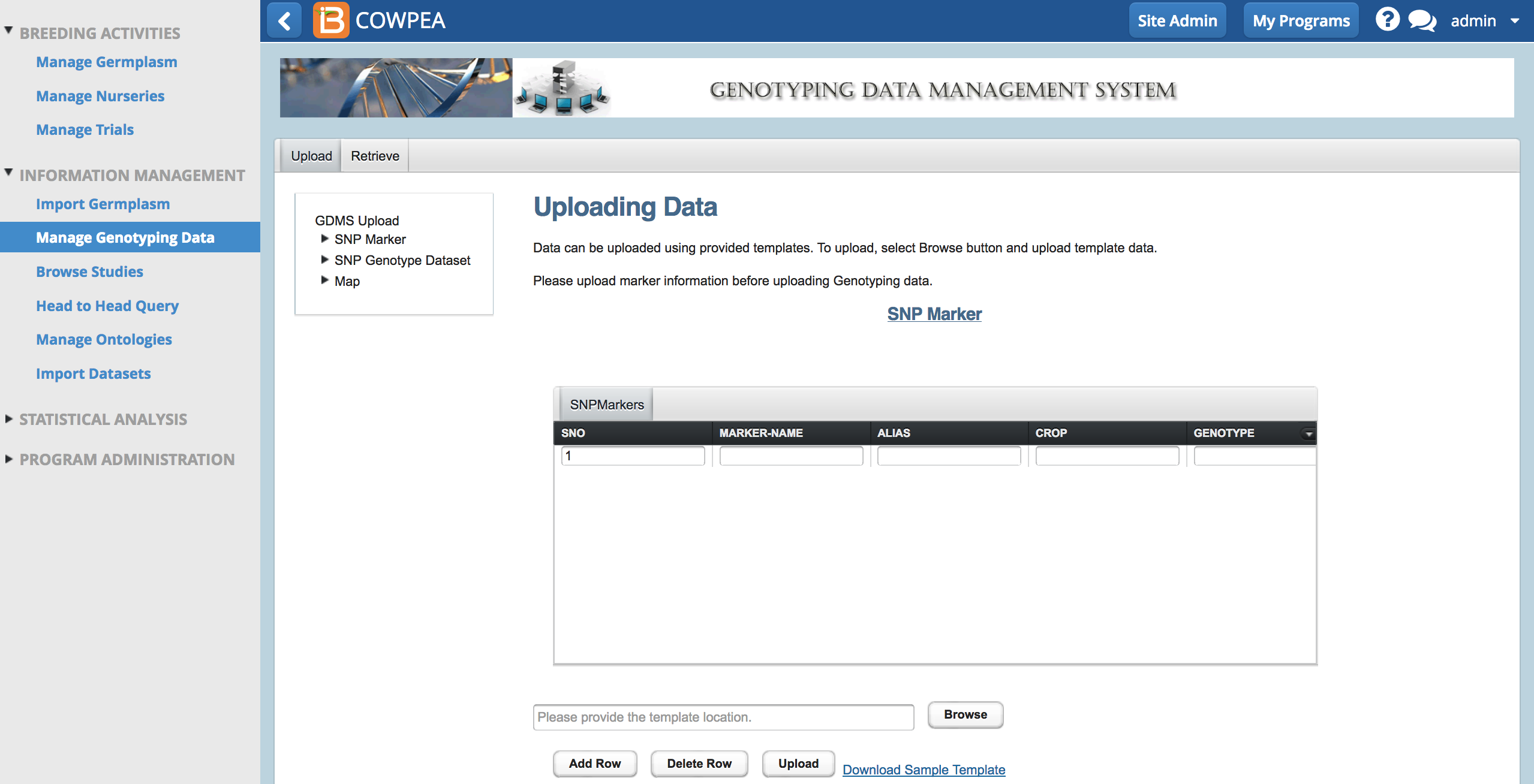
- Open the .xls file in Excel. Clear the sorghum example data from the .xls template.
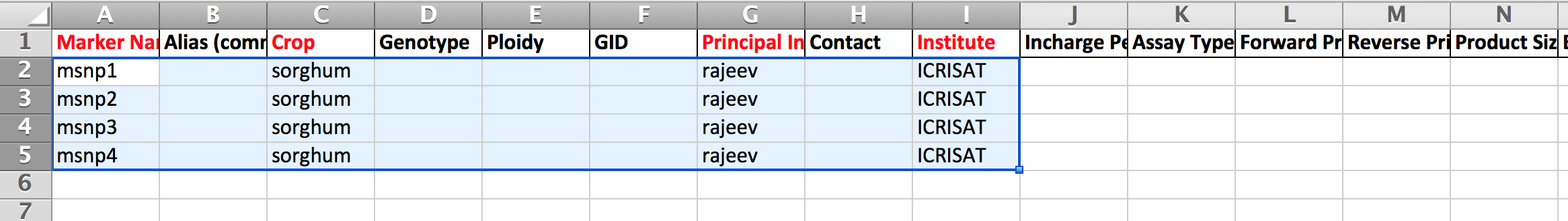
- Fill the mandatory red columns with marker information and save as .xls file. The black columns are for related optional marker data.
This example follows the upload of 2000 markers.
- Browse to the saved .xls file and select upload.
Retrieve
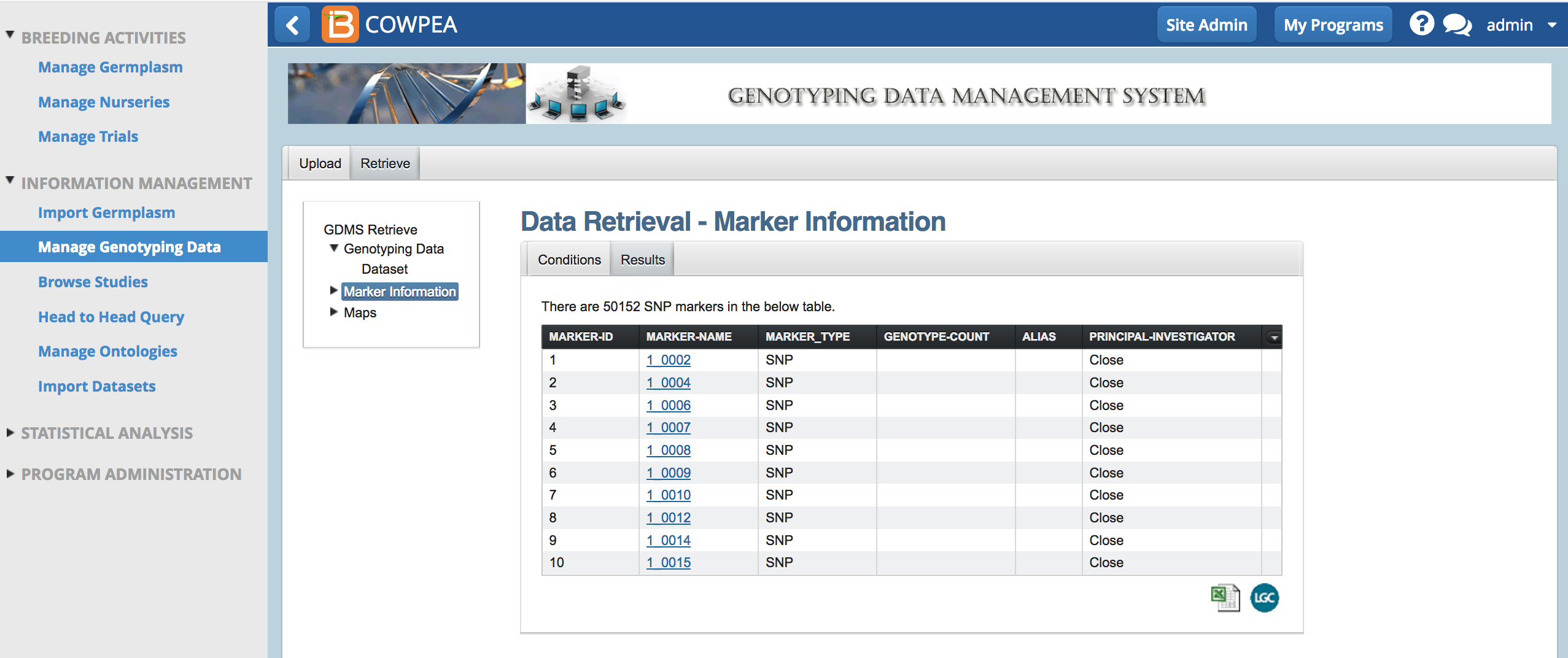
- Search for marker by name or retrieve all SNP markers.
- Export marker search results as an .xls file.
Genotype Data Sets
The BMS accepts genotype datasets in .csv format with columns corresponding to the markers and the rows to DNA samples. In BMS v9 DNA sampling is available from Manage Trials. Markers (see above) and samples (see Manage Samples) must be created in the system before sending for sequencing and uploading results.
Create Template
When samples are sent to a sequencing provider, like LGC, the sender must provide a unique id for all the samples and a list of markers to include in the genotyping assay.
- Download marker template (.xls) from Upload tab.
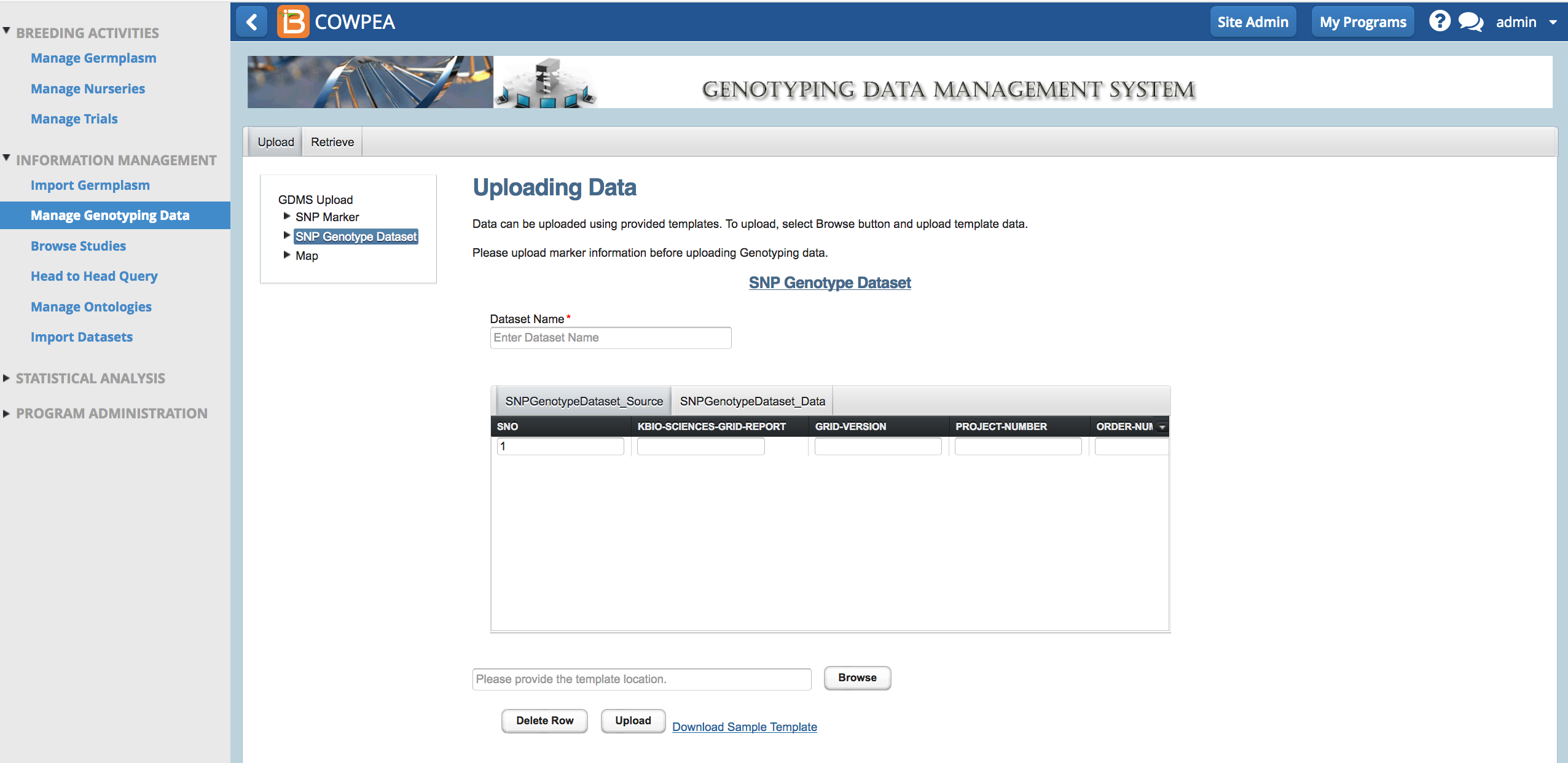
The sample template is formatted specifically for LGC genotyping services (previously KBioscence). The first 7 rows are optional for import back into the BMS. Rows 9-19 contain dummy sample data. Columns C-I contain dummy "MarkerName" data.
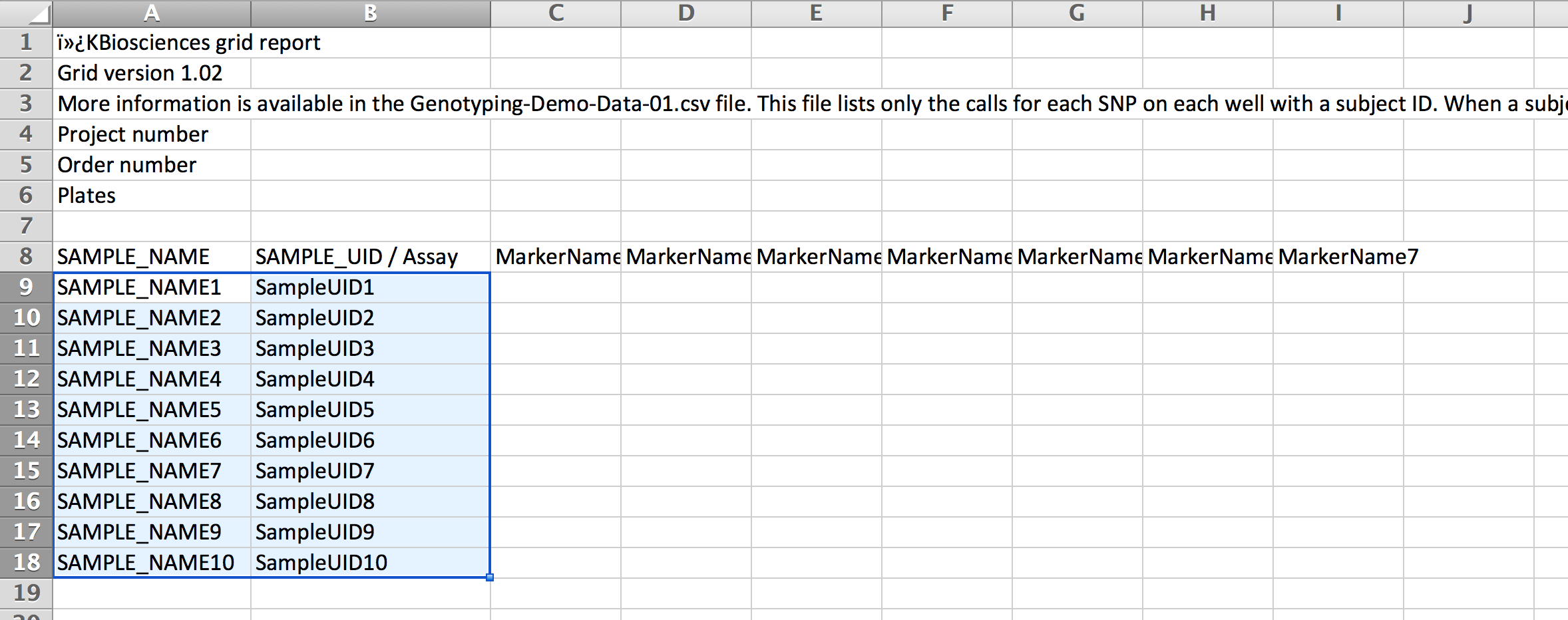
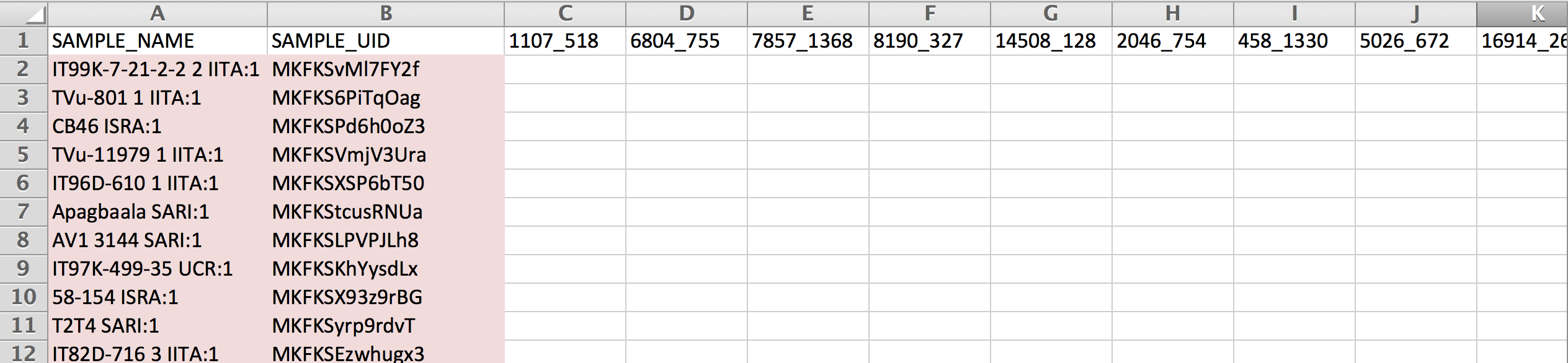
- Copy the SAMPLE_NAME and SAMPLE_UID from the sample list export file. Paste into the SAMPLE_NAME and SAMPLE_UID columns of the genotype template. Copy the marker names from the marker export file (see above). Save. This template is ready to send to a DNA sequencing service provider.
Upload Genotyping Results
Genotype service providers will send genotyping results (SNP calls) back in a matrix. To upload the data, the BMS needs the matrix in .csv format and will match the marker data to each SAMPLE_UID.
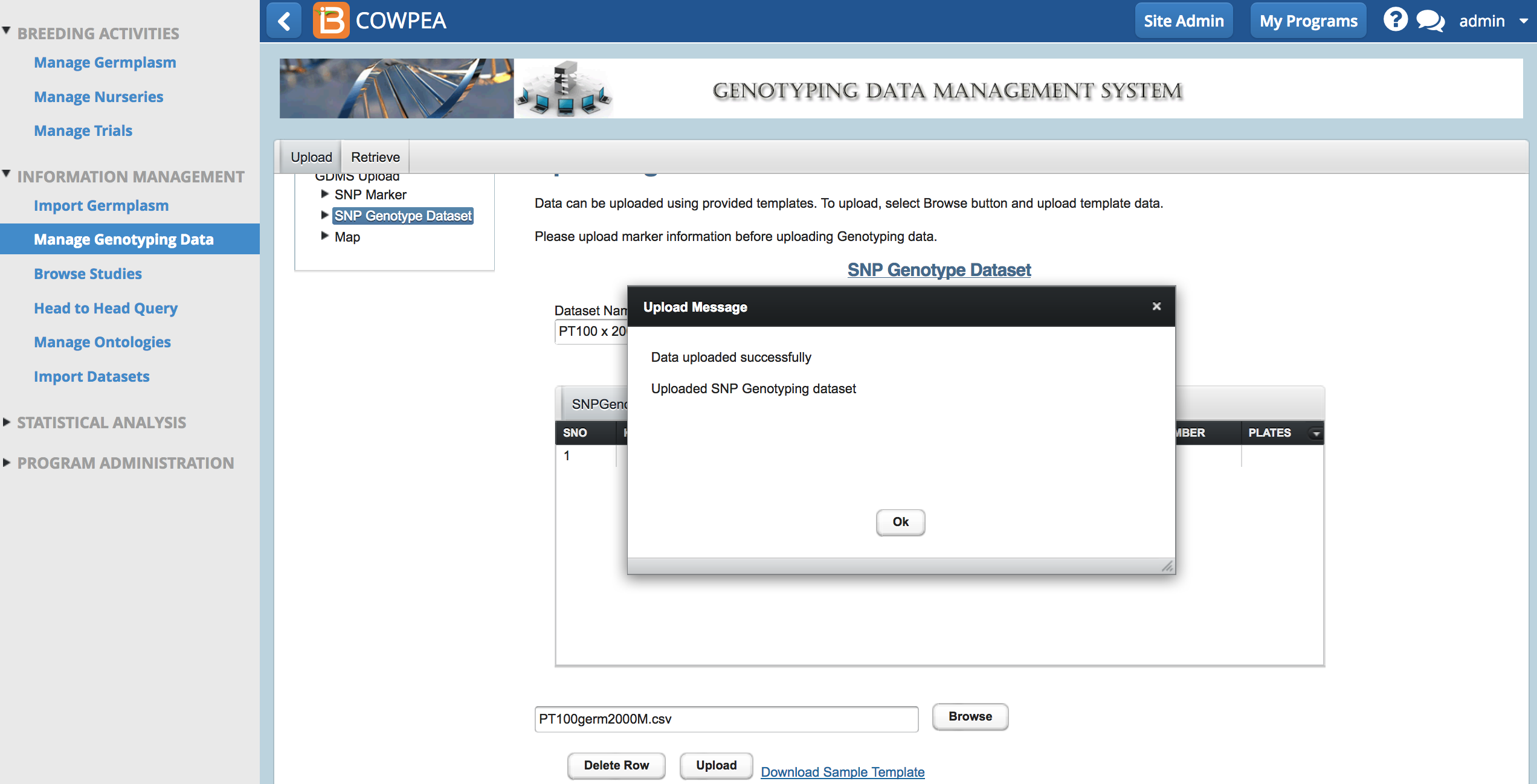
- From Upload SNP Genotype Dataset, name the data, browse to the .csv file, and select Upload.
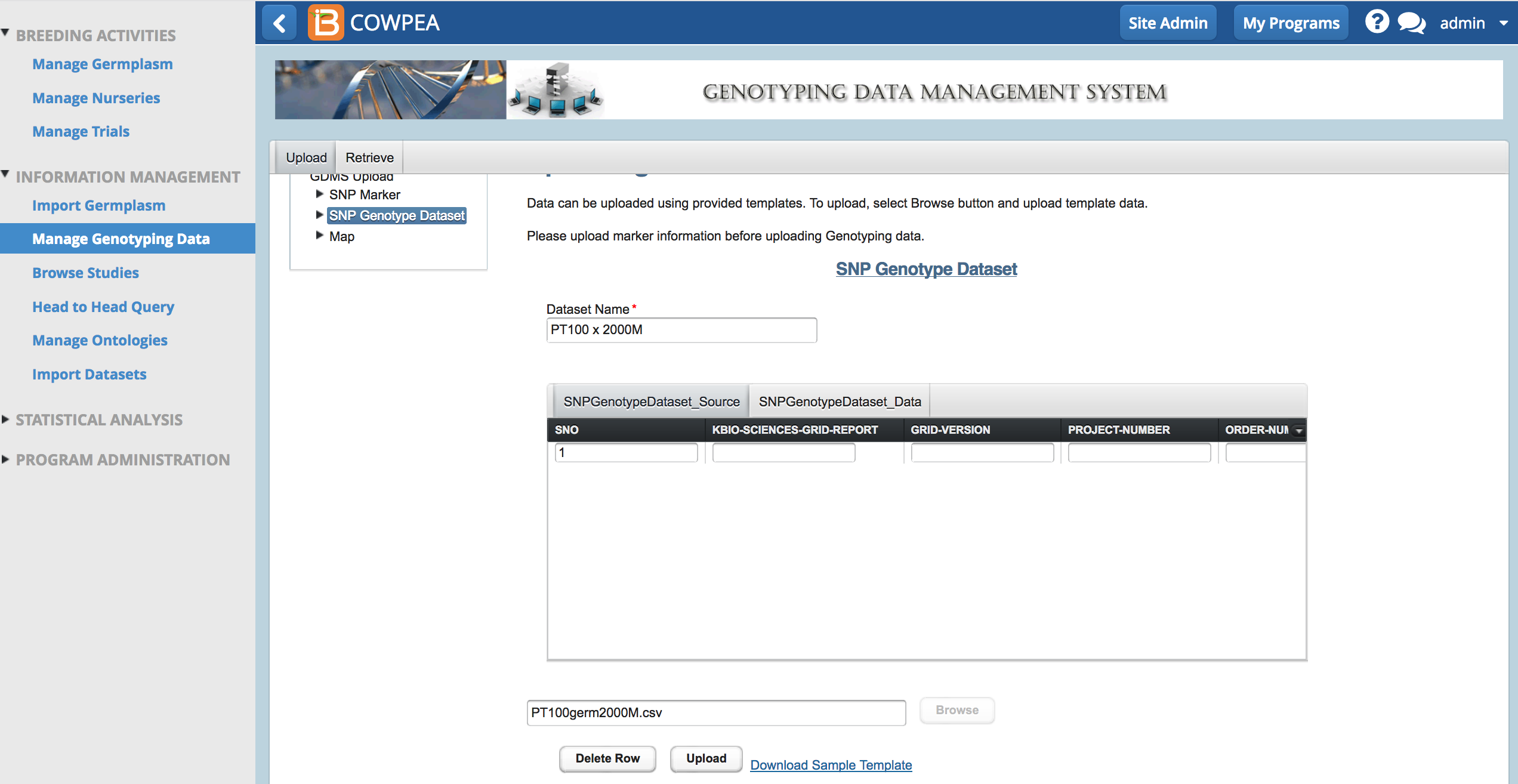
A success message appears when the dataset is uploaded. It may take several minutes to upload larger datasets. Data sets with more than 400,000 data points may freeze the system entirely and necessitate a server restart.
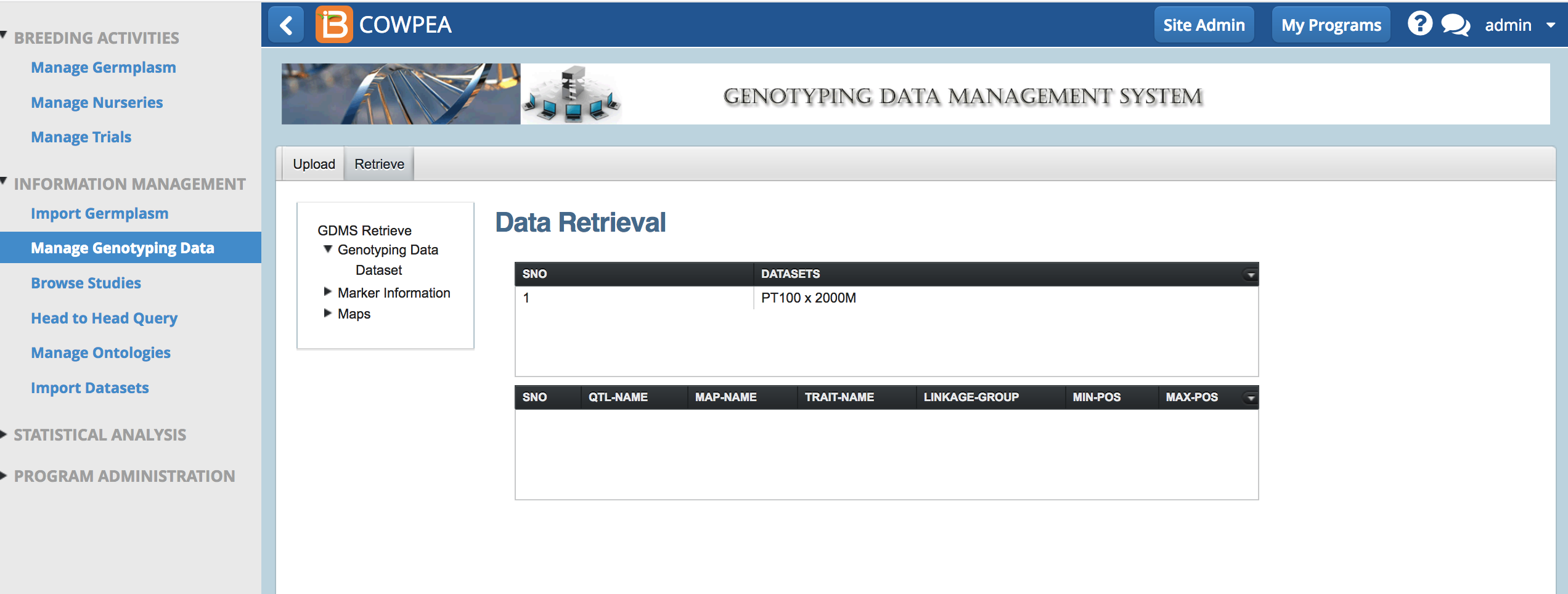
Retrieve Genotype Dataset
- Select the Retrieve tab and select Dataset.
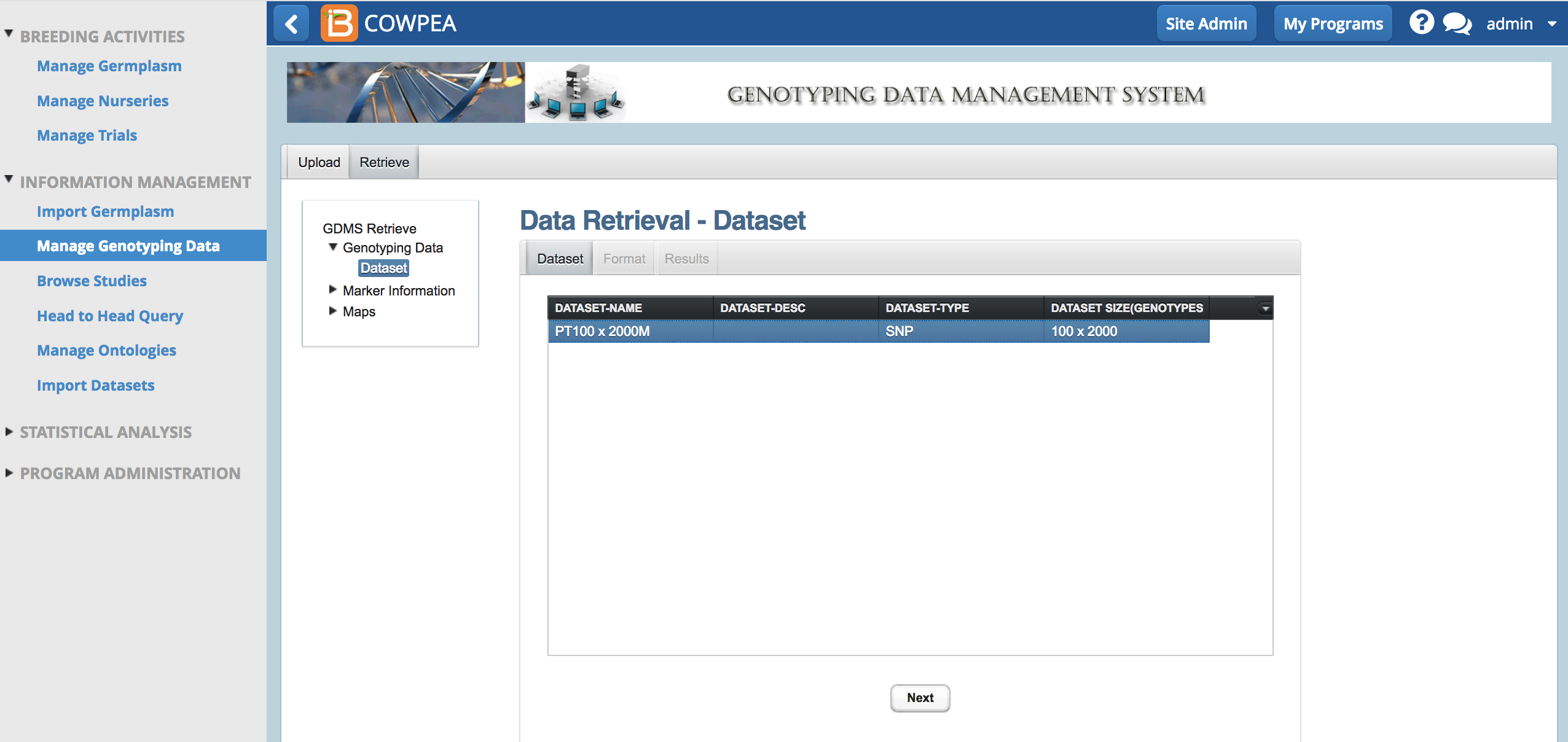
- Highlight the dataset of interest and select next.
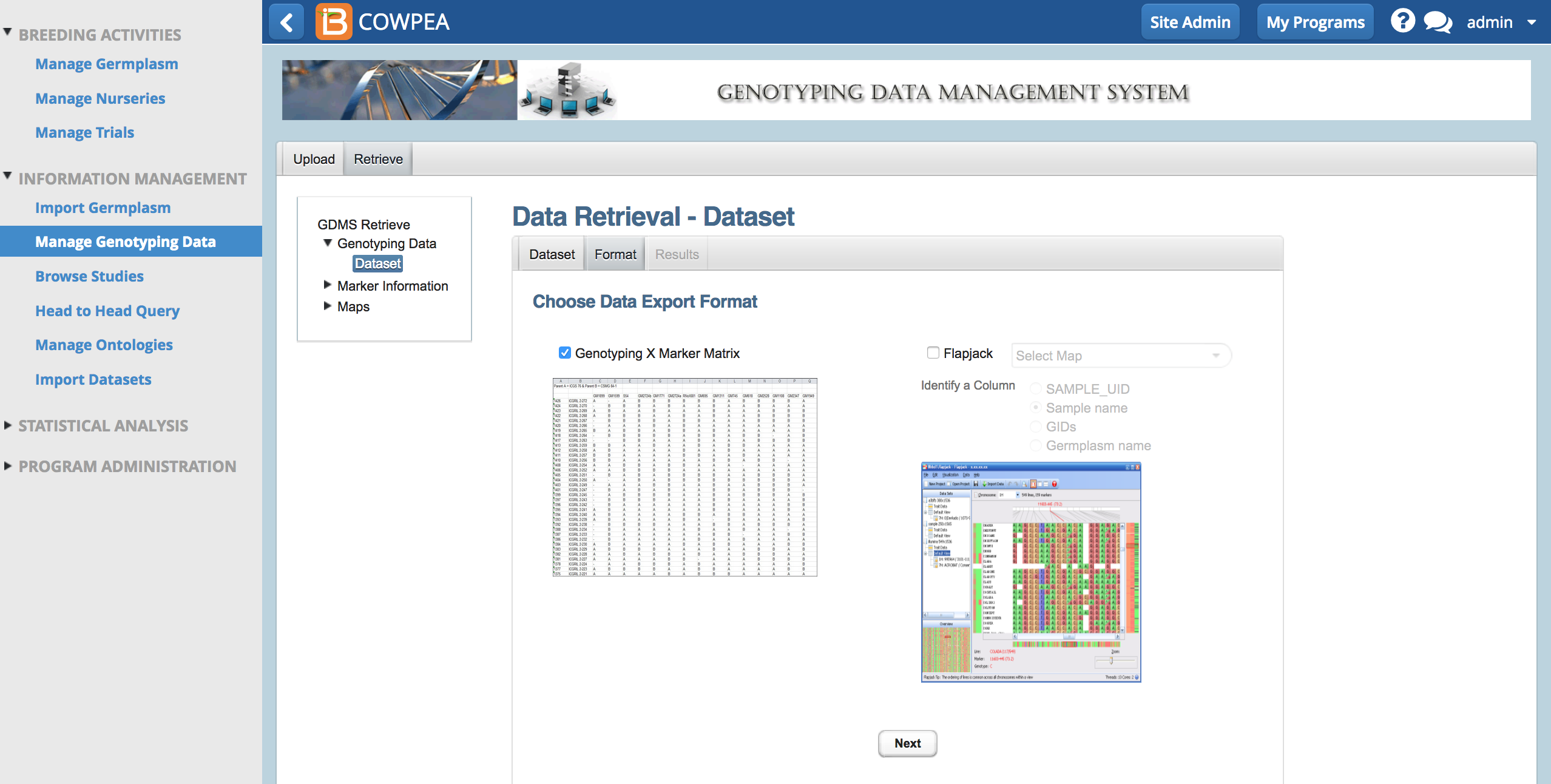
- Export the dataset as a genotype by marker matrix (.xsl) or as a Flapjack project files for visualization.
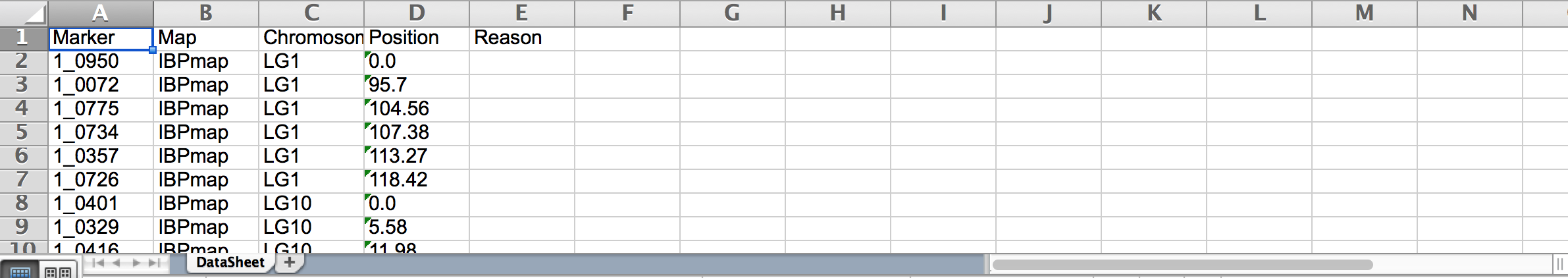
Maps
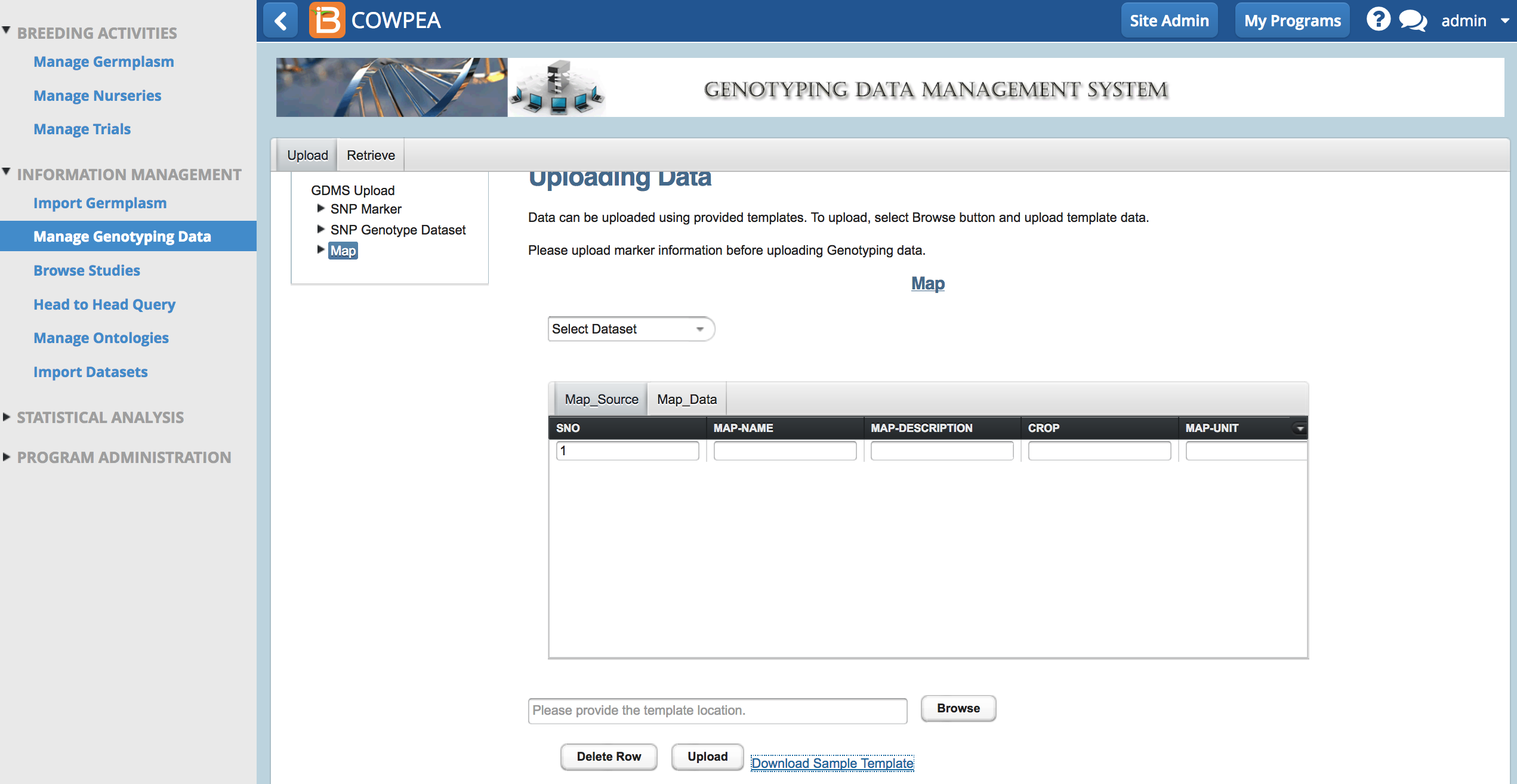
Upload
- Download sample template.
- Open the template (.xls) in Excel.
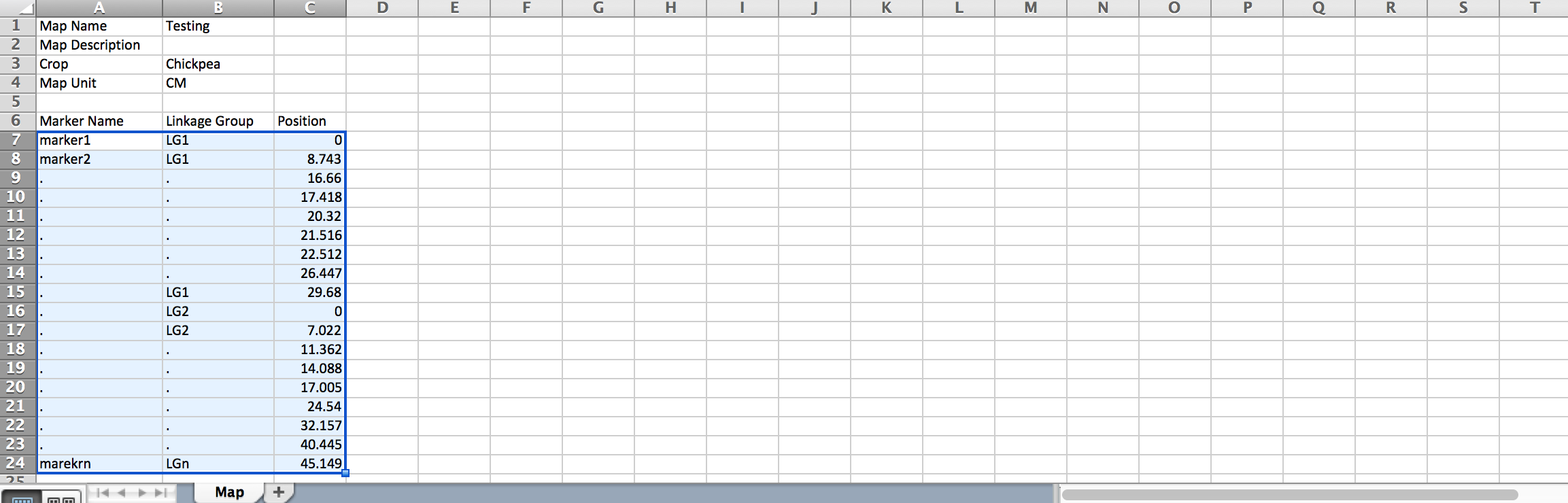
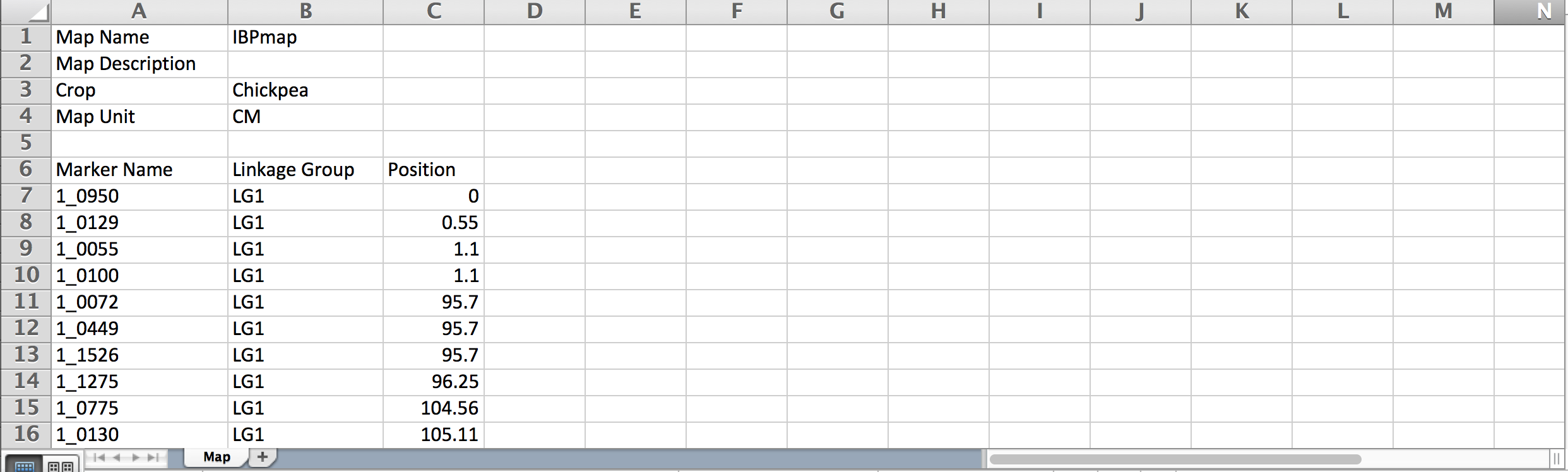
- Replace example data with actual map details. Save file.
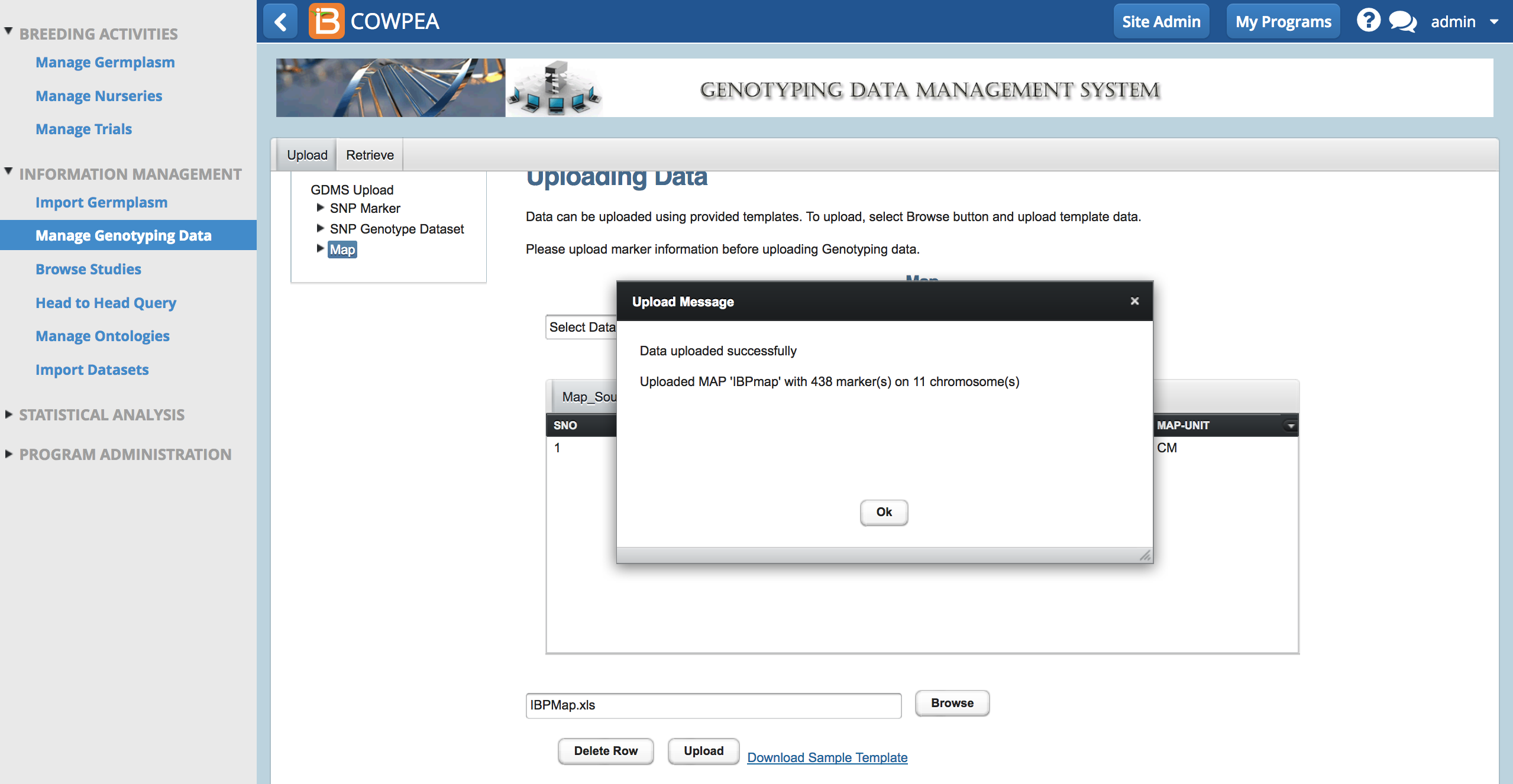
- Browse to the saved file. Select Upload.
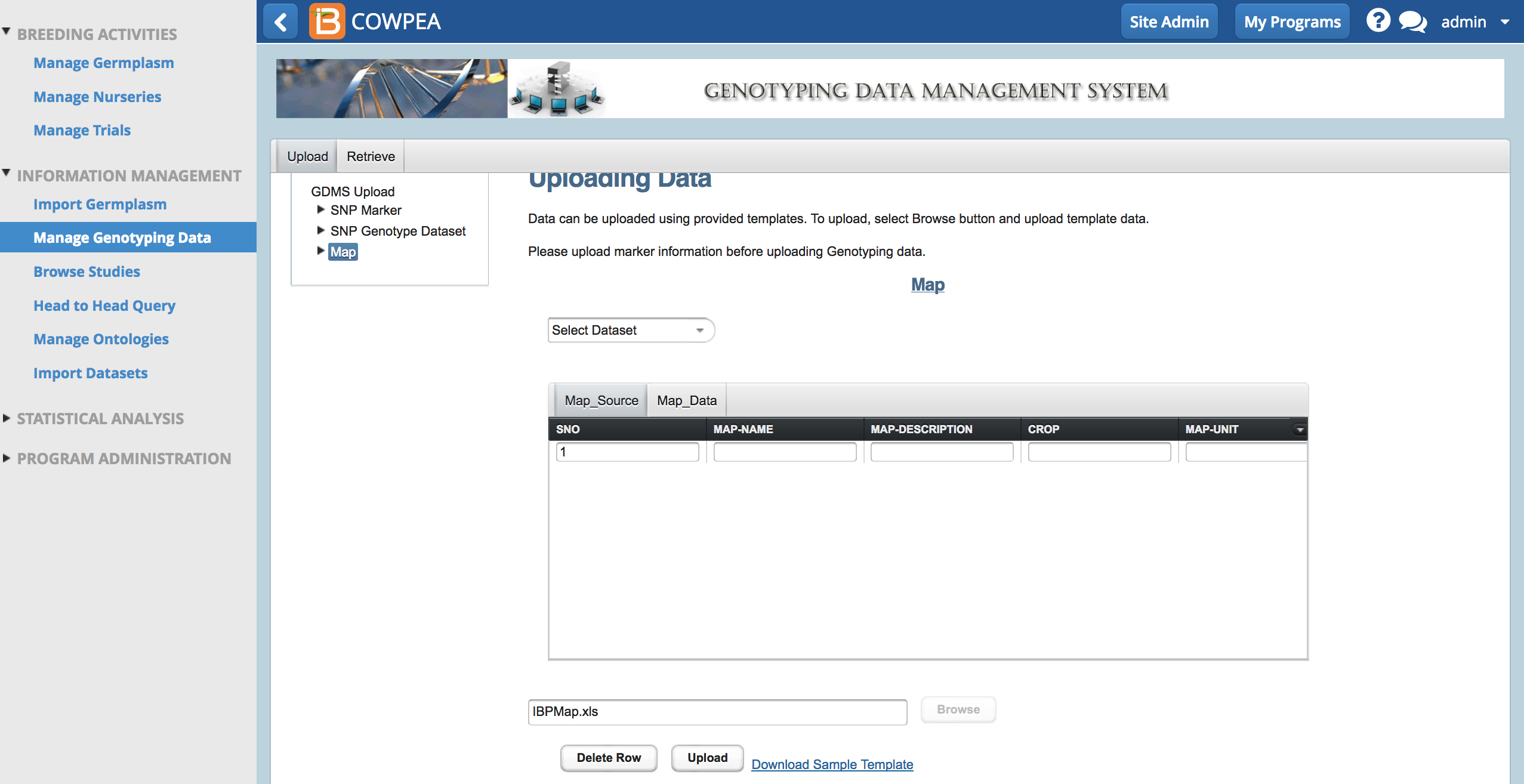
A success message appears when the map is uploaded.
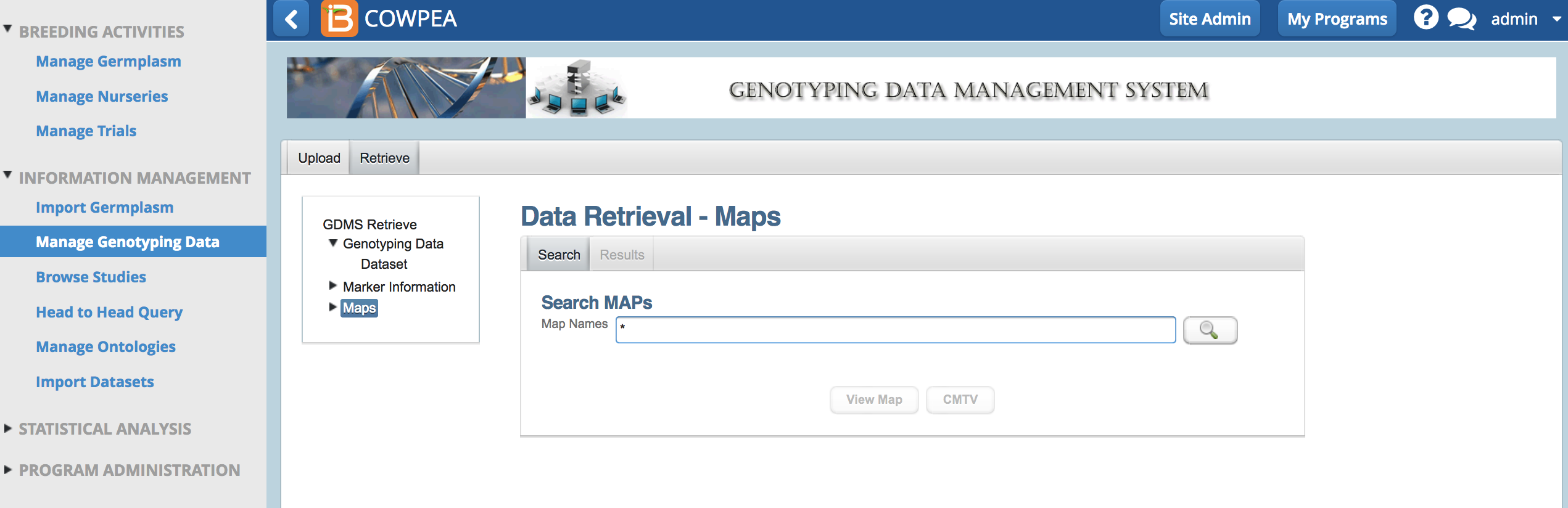
Retrieve
- From Retrieve Maps, search for map by name or enter asterisk * to retrieve all maps in the system.
- Select the map of interest and View Map.
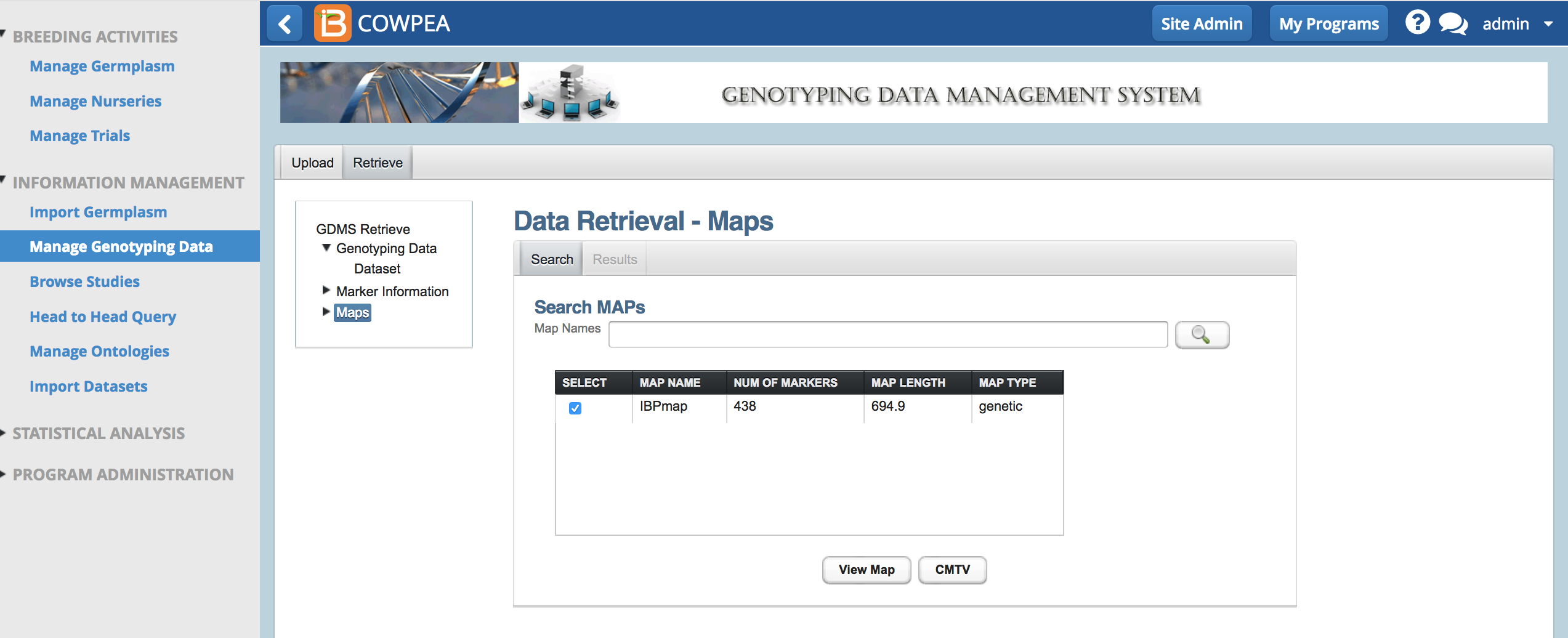
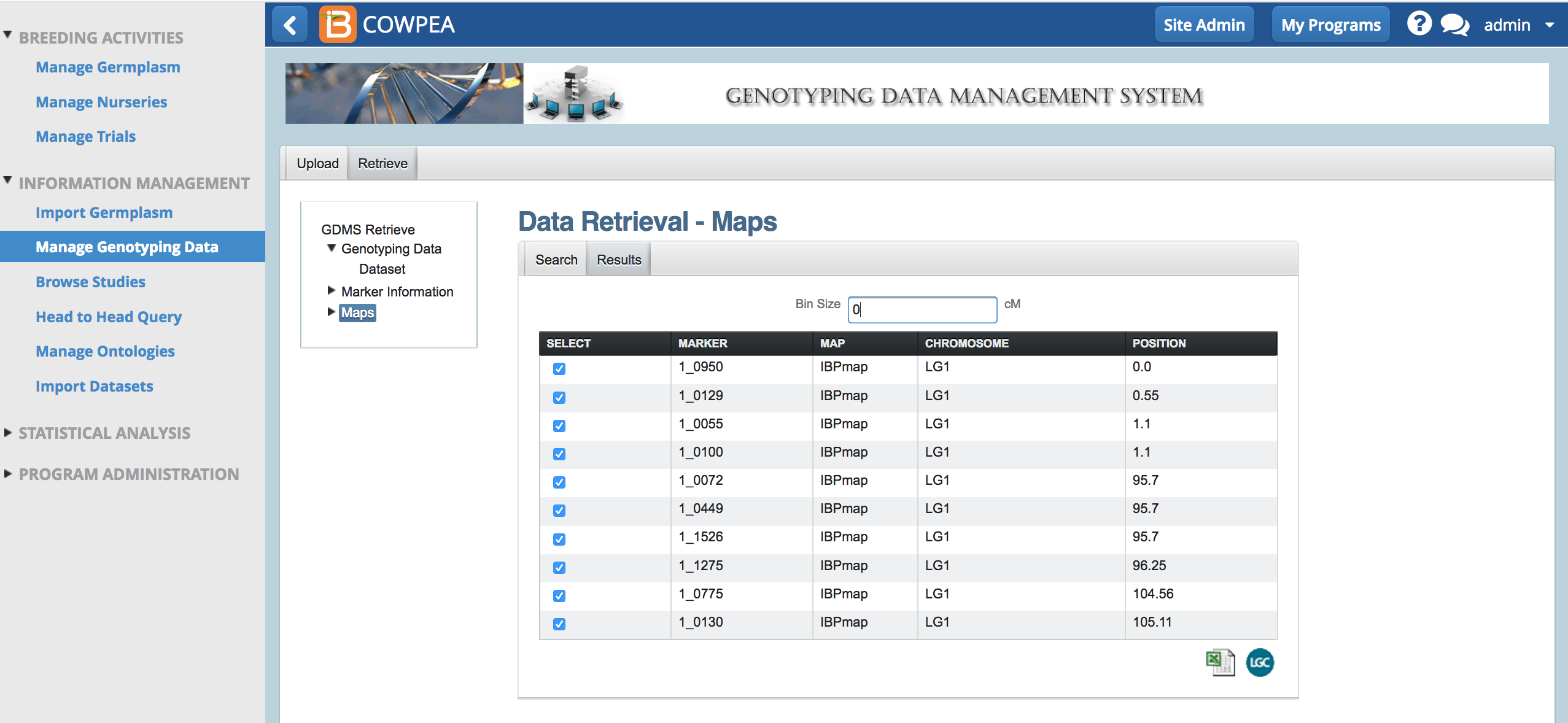
By default no markers are selected.
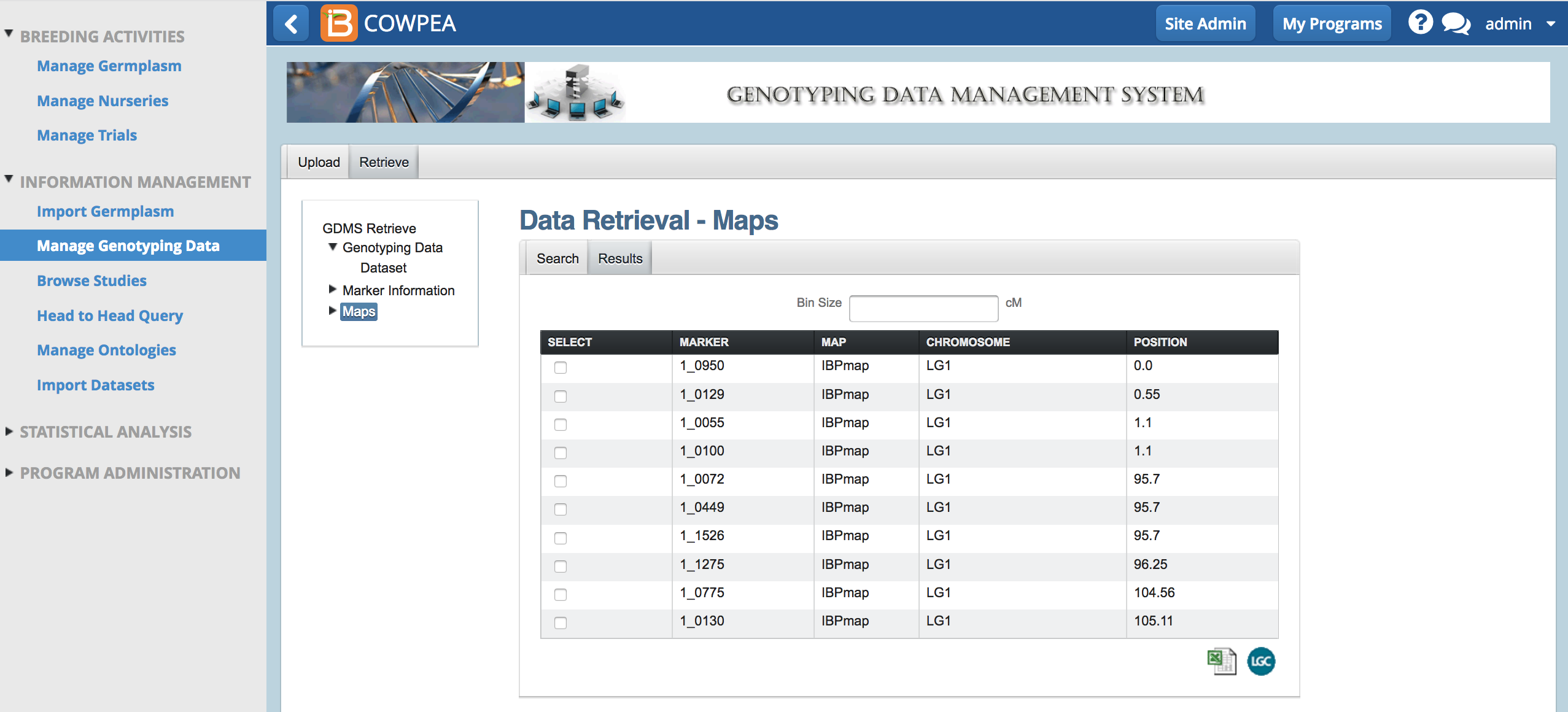
Marker Bin Size
- Select All markers by entering a bin size of zero. Export all the markers by selecting the Excel (.xls) option.
- Select fewer markers, equally across the genome, by entering bin sizes greater than zero. Export the markers in the bin by selecting the Excel option.
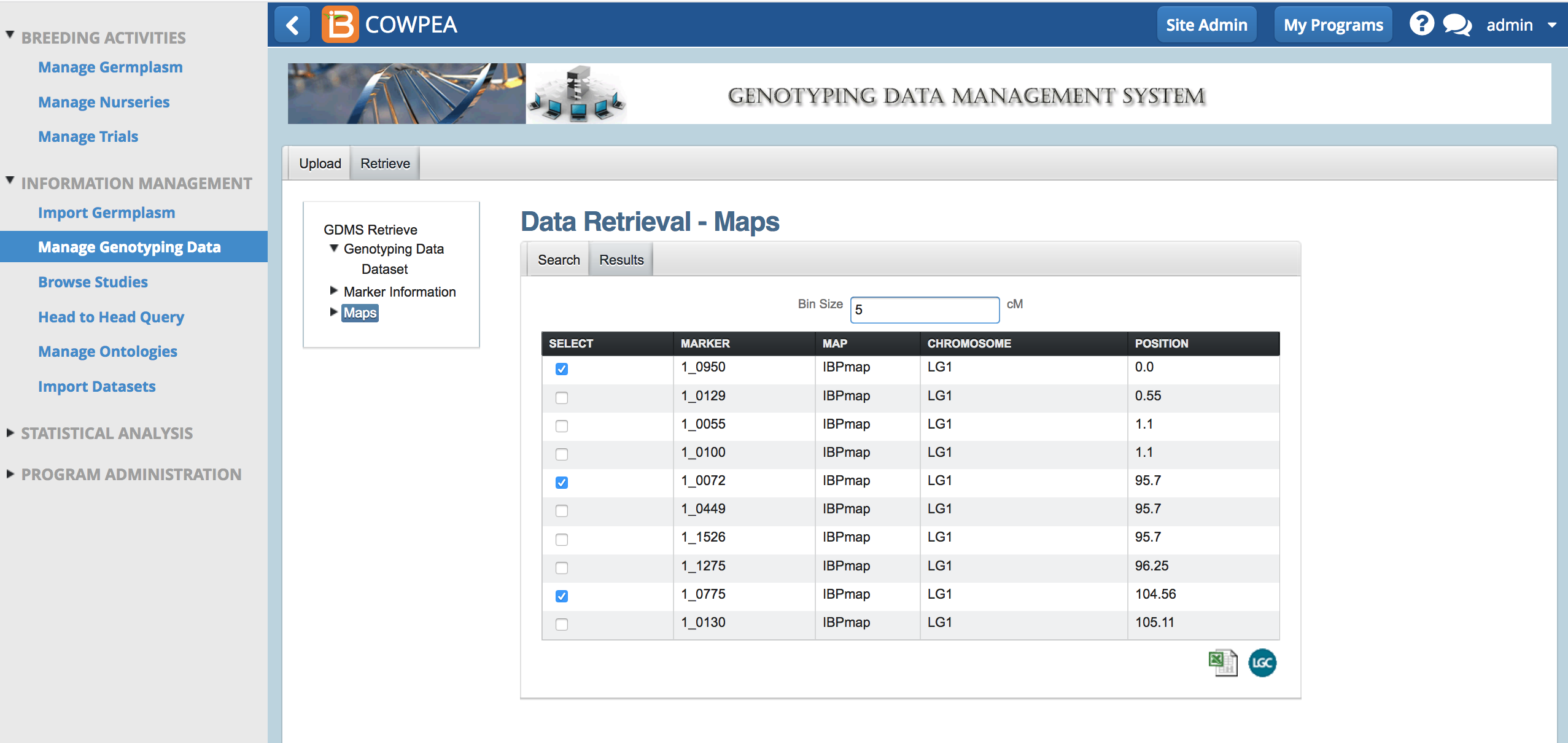
- Open the .xls file in Excel to review the exported markers.